December 11, 2017 weblog
DeepVariant: Tool to call out variants in sequencing data goes open source
(Tech Xplore)—Finding cats? That's old school. The AI that had been created by Google researchers Mark DePristo and Ryan Poplin built to recognize images of cats and dogs is, in Google Curiosity time, so yesterday. It's been a year and now they are on to using technology for finding gene mutations.
Their encouraging progress comes at a time when, as Will Knight wrote in MIT Technology Review, "making sense of the enormous amount of data that encodes human life remains a formidable challenge."
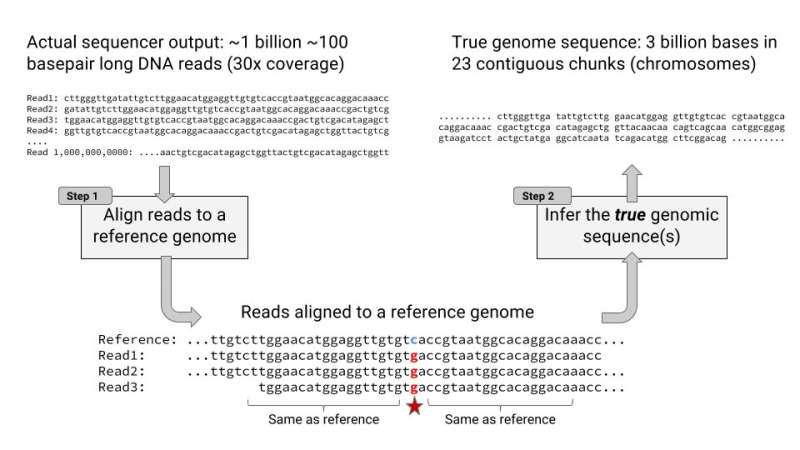
Megan Molteni, Wired, decoded, at least, the very nature of the challenge to know more about our human puzzle. "Today, a teaspoon of spit and a hundred bucks is all you need to get a snapshot of your DNA. But getting the full picture—all 3 billion base pairs of your genome—requires a much more laborious process. One that, even with the aid of sophisticated statistics, scientists still struggle over."
DeepVariant was developed by researchers from the Google Brain team, focused on AI techniques, and Verily, the Alphabet subsidiary focused on life sciences.
It is based on the same neural network for image recognition, but DeepVariant, is now making headlines not for cat IDs but as a way to scan a genetic code for mutations. DeepVariant has gone open source. The GitHub definition of DeepVariant: "an analysis pipeline that uses a deep neural network to call genetic variants from next-generation DNA sequencing data."
The researchers said it is deep learning technology with "significantly greater accuracy than previous classical methods."
Sophie Weiner, Popular Mechanics, said "it's better at recognizing gene mutations than any other program out there."
The FDA-administered 2016 PrecisionFDA Truth Challenge assessed several community-submitted variant callsets on the (at the time) blinded evaluation sample, HG002. DeepVariant won the Highest SNP Performance award.
One program already known is algorithm GATK, which used a lot of data in its attempt to figure out where sequencing may have gone wrong, said Weiner. DeepVariant is technically quite good at identifying mistakes in coding.
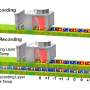
DeepVariant uses a different method to try to solve the glitches: "It turns the data into an image. Since Google's AI was originally used for image recognition, this technique ended up working really well."
Sarah Zhang in The Atlantic walked readers through the way in which DeepVariant works its magic without even knowing anything about DNA-sequencing machines.
"Neural networks are often analogized as layers of 'neurons' that deal in progressively more complex concepts—the first layer might respond to light, the second shapes, the third actual objects. As DeepVariant is trained with data, it learns which connections between 'neurons' to strengthen and which to ignore. Eventually, it can sort the actual mutations from the errors."
The task has turned visual. Zhang said, "The letters—A, T, C, or G—got assigned a red value; the quality of the sequencing at that location a green value; and which strand of DNA's two strands it is on a blue value. Together, they formed an RGB (red, green, blue) image."
DePristo was quoted in The Atlantic. "It changes the problem enormously from thinking super hard about the data to looking for more data."
Knight pointed out that it "automatically identifies small insertion and deletion mutations and single-base-pair mutations in sequencing data."
One thing GATK still has over DeepVariant as a tool for interpreting: speed. "The program functions at about half the speed of GATK," said Weiner.
Moving forward? "Programs like DeepVariant could use their complex data analysis abilities to predict the effects of a mutation, predicting which genes might activate," said Weiner. "The potential for the technology is unlimited, though we still have a way to go to catch up to the complexity of genes themselves."
In a Dec. 4 Google Research Blog, team members stated DeepVariant's release as open source was to accelerate the use of this technology to solve real-world problems.
"To further this goal, we partnered with Google Cloud Platform (GCP) to deploy DeepVariant workflows on GCP, available today, in configurations optimized for low-cost and fast turnarounds using scalable GCP technologies like the Pipelines API. This paired set of releases provides a smooth ramp for users to explore and evaluate the capabilities of DeepVariant in their current compute environment."
They said it also provided a scalable, cloud-based solution to satisfy needs of even the largest genomics datasets.
More information:
github.com/google/deepvariant
research.googleblog.com/2017/1 … ccurate-genomes.html
© 2017 Tech Xplore