New artificial neuronal networks can now recognize and assign nerve cells independently based on their appearance. Credit: Max Planck Society
Is it possible to understand the brain? Science is still far from answering this question. However, since researchers have started training artificial intelligence on neurobiological analyses, it seems at least possible to reconstruct the cellular structure of a brain. New artificial neural networks developed by the Max Planck Institute of Neurobiology and Google AI can now even recognize and classify nerve cells independently based on their appearance.
The human brain consists of about 86 billion nerve cells and about as many glial cells. In addition, there are about 100 trillion connections between the nerve cells alone. While mapping all the connections of a human brain remains out of reach, scientists have started to address the problem on a smaller scale. Through the development of serial block-face scanning electron microscopy, all cells and connections of a particular brain area can now be automatically surveyed and displayed in a three-dimensional image.
"It can take several months to survey a 0.3 mm3 piece of brain under an electron microscope," says Philipp Schubert, doctoral student in Winfried Denk's Department at the Max Planck Institute of Neurobiology. "Depending on the size of the brain, this seems like a lot of time for a tiny piece. But even this contains thousands of cells." Such a data set would also require almost 100 terabytes of storage space. However, it is not the collection and storage but rather the data analysis that is the difficult part.
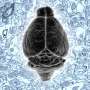
Philipp Schubert studied computational physics at the University of Heidelberg. Since 2017, he has been developing artificial neural networks as part of his doctoral thesis at the Max Planck Institute of Neurobiology with the aim of largely automating connectome analysis. The data is collected at the serial block-face scanning electron microscope shown here. Credit: Max Planck Society
Fortunately, the methods of analysis have improved alongside the microscopy techniques. For a long time, only the human brain seemed to be able to reliably recognize and track the parts and connections of nerve cells in electron microscope images. For example, people still had to work for hours in front of a computer screen to track cell components in the image stacks and correct computer analyses. As a result, the reconstruction of even the smallest data sets took many years. A few years ago, however, the researchers enlisted the help of artificial intelligence.
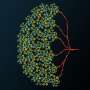
The neurobiologists from Martinsried trained convolutional neural networks to recognize and distinguish nerve cell components in the image data. By means of improved image analysis using flood filling networks, entire nerve cells with all their components and connections were automatically extracted from an image stack in 2018—with virtually no errors. "And now, with the cellular morphology neural networks, we are going one step further in the analysis," says Schubert. "Like humans, CMNs recognize a cell by its shape and context and not by comparing individual pixels."
The CMNs can now assign the nerve cells extracted from an image stack to a nerve cell type or a glial cell depending on their appearance. The CMNs also recognize whether a cell area belongs to the cell body, the axon, a dendrite, or its spinous processes. "This information is important in order to be able to understand the function of the cells or, for example, the direction of the information flow at the synaptic contact points, says Schubert, who is already looking forward to the next task: "Now we can finally analyze the larger data sets!"
More information: Philipp J. Schubert et al. Learning cellular morphology with neural networks, Nature Communications (2019). DOI: 10.1038/s41467-019-10836-3
Journal information: Nature Communications
Provided by Max Planck Society