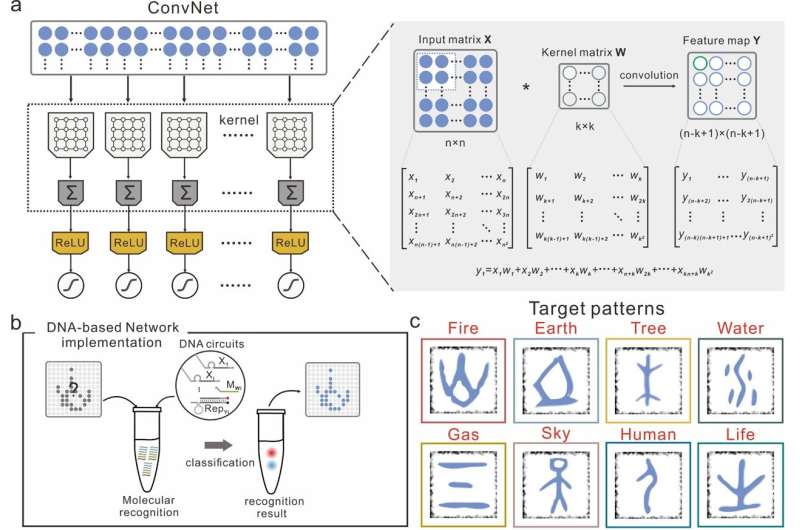
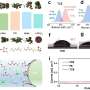
Molecular convolutional neural networks with DNA regulatory circuits. (a) Left: The architecture of ConvNet. Right: Schematic of the operation principle of ConvNet for recognition tasks. (b) Unknow input pattern is added to the solution, upon addition of the DNA circuit, fluorescence signals can be read out to report recognition results. (c) The DNA-ConvNet can recognize eight Chinese oracles. Credit: Xiong et al.
Molecular computing is a promising area of study aimed at using biological molecules to create programmable devices. This idea was first introduced in the mid-1990s and has since been realized by several computer scientists and physicists worldwide.
Researchers at East China Normal University and Shanghai Jiao Tong University have recently developed molecular convolutional neural networks (CNNs) based on synthetic DNA regulatory circuits. Their approach, introduced in a paper published in Nature Machine Intelligence, overcomes some of the challenges typically encountered when creating efficient artificial neural networks based on molecular components.
"The intersection of computer science and molecular biology is a fertile ground for new and exciting science, especially the design of intelligent systems is a longstanding goal for scientists," Hao Pei, one of the researchers who carried out the study, told TechXplore. "Compared to the brain, the scale and computing power of developed DNA neural networks are severely limited, due to the network size limitations. The primary objective of our work was to scale up the computing power of DNA circuits by introducing a suitable neural network model for DNA molecular systems."
While conducting their research, Pei and his colleagues found that could be particularly promising for modeling DNA circuits, due to their sparse topological connectivity, which resembles that of real biological neural networks. They thus decided to use CNNs to devise a large-scale DNA-based neural network comprised of 512 synthetic DNA strands. Notably, their proposed network produces thousands of chemical reactions and generates hundreds of molecular species.
"Our group has been focused on the precisely engineering and programming of nucleic acids molecules, and we have designed and constructed a series of dynamic DNA nanostructures that could potentially be used as regulatory elements for the construction of large circuits," Pei explained. "In this work, we make use of a dynamic DNA nanostructure called switching gate, which is functionally similar to the riboswitches in gene regulatory circuits, all consisting of two independent functional domains that sense and respond to external inputs."
The switching gate in the researchers' network allows them to independently control its signal transmission functions and weight assignment functions through a process known as intramolecular conformational switching. This process is particularly suited for the weight-sharing and sparse connectivity of CNNs.
The DNA circuits operate within the researchers' network, where all computing units are ready to respond to inputs. Once the inputs are fed to the solution, single DNA strands will trigger cascaded strand displacement reactions in order.
These reactions, driven by the Gibbs free energy or entropy in the system, generate corresponding fluorescent signals. The researchers' encoded all test patterns using a set of single DNA strands and each of the generated fluorescence signals represents one of these test patterns.
"We have extended the key feature of the CNN―sparse topology and weight-sharing architecture to a DNA neural network, which can effectively reduce complexity and parameters of network architecture through sparsely connected neurons," Pei said. "To implement these features, we have designed switching gate architecture composed of two independent functional domains (weight tuning domain and recognition domain). With this design, we can easily change the sequence design of corresponding functional domain respectively to fit the adjust of network architecture."
The CNN-based approach has several advantages over previously proposed molecular computing methods. Firstly, its switching gate architecture could be used to embed ligand-responsive molecular switches. This would allow the network to adapt its functions in response to environmental changes, potentially enabling the development of molecular circuits that resemble biological neural networks and capable of "intelligent" behavior.
In addition, the inherently parallel nature of DNA molecules could enable the autonomous parallelization of the CNN's operations. This could be particularly valuable for achieving scalable information processing.
"We have proposed a systematic strategy for implementing the ConvNet algorithm at the molecular level," Pei said. "We feel that our method is a major advancement in artificial molecular information-processing systems, as it achieved rapid and accurate classification tasks that could classify 32 molecular patterns within 30 min, which might be the fastest and the most powerful and complex artificial chemical computing system to date, as far as we know."
The recent work by Pei and his colleagues introduces an alternative DNA-based architecture that could inform the design of new molecular computing systems. In the future, their approach could be used to create various molecular diagnostic devices for biomedical applications.
"By interfacing sensory inputs, the DNA-based ConvNet could in principle use hundreds of targets as inputs and facilitate broader applications in disease diagnostics, profiling expression patterns, and precision medicine," Pei added. "Based on this DNA-based ConvNet model, we now plan to construct molecular classifier that can be used for multi-disease diagnostic classification."
More information: Xiewei Xiong et al, Molecular convolutional neural networks with DNA regulatory circuits, Nature Machine Intelligence (2022). DOI: 10.1038/s42256-022-00502-7
Wei Lai et al, Programming Chemical Reaction Networks Using Intramolecular Conformational Motions of DNA, ACS Nano (2018). DOI: 10.1021/acsnano.8b02864
Wei Lai et al, Nonlinear Regulation of Enzyme-Free DNA Circuitry with Ultrasensitive Switches, ACS Synthetic Biology (2019). DOI: 10.1021/acssynbio.9b00208
Journal information: Nature Machine Intelligence , ACS Nano , ACS Synthetic Biology
© 2022 Science X Network