This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Unleashing the power of soft-decision decoding in DNA digital storage
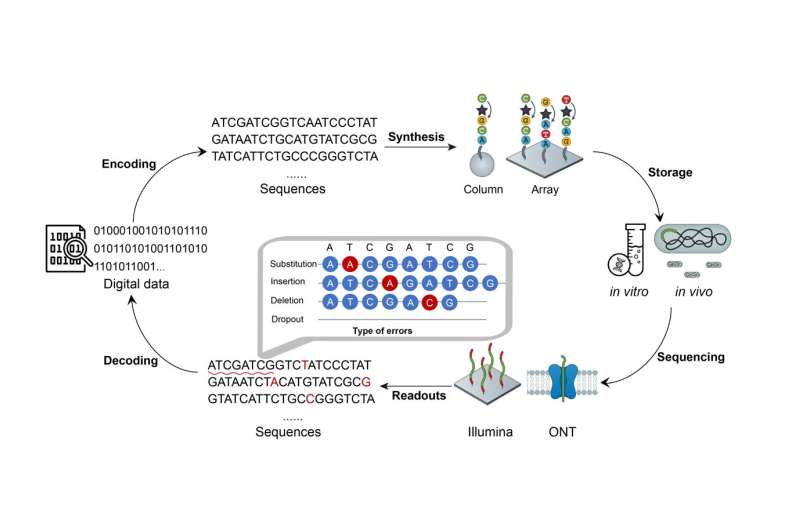
Led by Dr. Jue Ruan and Dr. Weihua Pan, a study published in the journal National Science Review delves into the realm of DNA digital storage (DDS), a technology acclaimed for its high-density (EB/g), long-term (million years) and low maintenance costs, offering a promising solution for the ever-growing demands of big data storage.
A key challenge in DDS lies in the high error rates, which pose difficulties in data recovery and compromise storage density due to the redundancy added for error correction. "Through an in-depth analysis of error correction principles, we were thrilled to discover soft-decision decoding, a technique used in the communication field to predict and correct errors without sacrificing information density," says Dr. Ding, the co-first author of the research paper.
However, unlike binary sequences in communication engineering, DDS involves four nucleotides and various error types, including substitutions, insertions, and deletions, thereby challenging error prediction. The team addressed this by developing an accurate error prediction model based on the analysis of the DDS process, sequencing data, and alignment.
"We initially don't know the number of errors, so we provide a large candidate set for error prediction. By iterating the candidate set with error correction techniques, we can achieve successful error correction only when the prediction accurately identifies enough number of errors," explains Wu, the co-first author of the research paper.
To ensure accurate recovery of information, Derrick incorporates a checksums algorithm for secondary verification of error-corrected data. Additionally, a backtracking algorithm enables error identification and re-decoding upon checksum algorithm detecting errors.
Through error prediction, error correction and implementing soft-decision decoding, Derrick surpasses the limitations of error-correcting abilities in hard-decision decoding, theoretically extending the upper limit of error correction to infinity. In practical applications, Derrick successfully recovers MB-level file data with 100% accuracy, doubles the error-correcting capability of Reed-Solomon code, and achieves the optimal balance between error correction overhead and storage density in the field.
Moreover, this research presents a fundamental improvement in error correction techniques applied to DNA digital storage. Previous studies in the field can significantly benefit from incorporating the newly introduced soft-decision strategy, leading to a substantial enhancement in error correction capabilities.
More information: Lulu Ding et al, Improving error-correcting capability in DNA digital storage via soft-decision decoding, National Science Review (2023). DOI: 10.1093/nsr/nwad229