July 23, 2018 feature
Developing brain atlas using deep learning algorithms
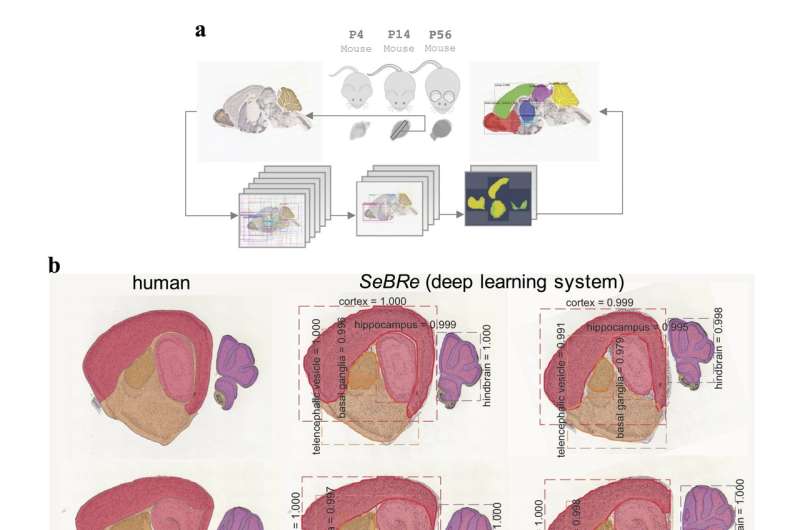
A team of researchers from the Brain Research Institute of the University of Zurich and the Swiss Federal Institute of Technology (ETH) have developed a fully automated brain registration method that could be used to segment brain regions of interest in mice.
Neuroscientists are always seeking out new methods of exploring the structure and function of different brain regions, which are initially applied on animals but could eventually lead to important discoveries about the organization of the human brain.
"My lab aims to reveal how the mammalian brain develops its abilities to process and react to sensory stimuli," Theofanis Karayannis, one of the researchers who carried out the study told Tech Xplore. "Most of the work we do is on the experimental side, utilizing the mouse as a model system and techniques that range from molecular-genetic to functional and anatomical."
This study is part of a larger project, which also includes "Exploring Brain-wide Development of Inhibition through Deep Learning," a study in which Karayannis and his colleagues use deep learning algorithms to comprehensively track the so-called inhibitory neurons over time in order to gauge the development of capabilities of the brain at specific points in time.
To do that, they tried to devise a method that could accurately delineate and segment different brain regions in experimental images of the developing mouse brain, to then extract information about the location and density of inhibitory neurons.
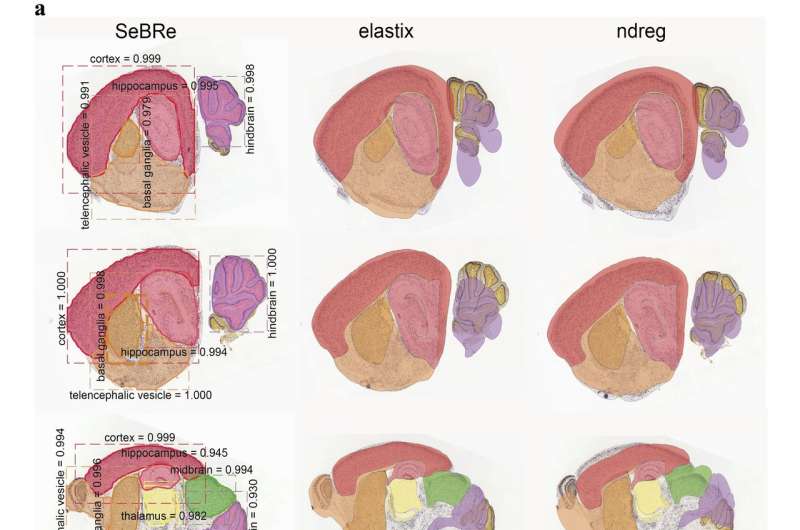
"By utilizing the computational skills of Asim Iqbal, a PhD student in my lab, we sought out to first test the utility of a few image-registration based methods that have been gaining attention in neuroscience studies over the last year," explains Karayannis. "We quickly realized that existing techniques are suboptimal for cases where the tissue sections are rotated or when their geometry is compromised due to methodological issues, for example during brain tissue slicing."
After observing the limitations of existing image registration-based methods, the researchers set out to develop a new deep learning tool that can produce reliable results regardless of possible scale, rotation and morphological issues affecting sections of brain tissues.
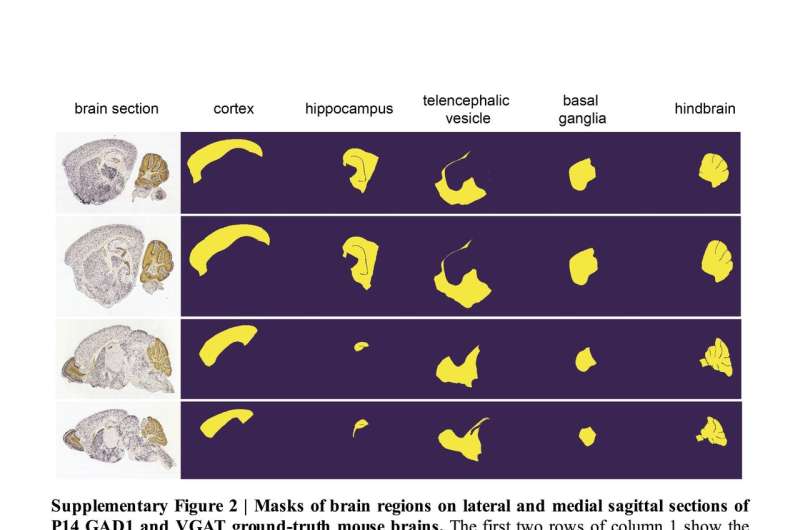
This method, called SeBRe (Segmenting Brain Regions), allows for registration through segmenting brain regions of interest, which could assist scientists in their studies of brain regions across a variety of development stages. SeBRe takes brain sections, as well as the binary masks of brain regions, as input for training.
The researchers trained their neural network on brain sections of 14 days old mice, for two genetic markers. They then tested its performance in generating anatomical masks of previously unidentified sections of the brain of 4, 14, 28, and 56 days old mice, across a range of neuronal markers. SeBRe outperformed all existing brain registration methods, providing the minimum mean squared error (MSE) score on a mouse brain dataset.
"Our study provides a novel, robust approach to the current affine and non-affine methods for brain area registration," says Karayannis. "It also points at the applicability of an artificial intelligence-based method in segmenting brain structures of interest."
In future, SeBRe could be instrumental in tracking and quantifying anatomical changes in the brain during development, as well as identifying other meaningful information, such as what genes are expressed during the development of a mouse brain and their spatio-temporal qualities.
So far, their deep learning algorithm has only been trained on a few brain regions and the researchers found that it performed sub-optimally on other areas with complex 3-D structures.
Karayannis and his colleagues are now planning to scale their method to segment various sub-regions in the mouse and human brain, by using more annotated data to further optimize the performance of their algorithm.
"Hopefully, this study is the beginning of a path that will ultimately allow us and the community to explore alterations in brain structure and function, not only across different developmental stages, but also in devastating brain disorders, helping to identify new biomarkers and form novel hypotheses about disease generation and progression," says Karayannis.
More information: Developing Brain Atlas through Deep Learning, arXiv:1807.03440 [cs.CV] arxiv.org/abs/1807.03440
Exploring Brain-wide Development of Inhibition through Deep Learning, arXiv:1807.03238 [cs.CV] arxiv.org/abs/1807.03238
© 2018 Tech Xplore