Reconstructing histological slices into 3-D images
Despite advances in 3D imaging such as MRI and CT, scientists still rely on slicing a specimen into 2-D sections to acquire the most detailed information. Using this information, they then try to reconstruct a 3-D image of the specimen. Researchers from Nara Institute of Science and Technology report a new algorithm that can do this task at less cost and higher robustness than standard methods.
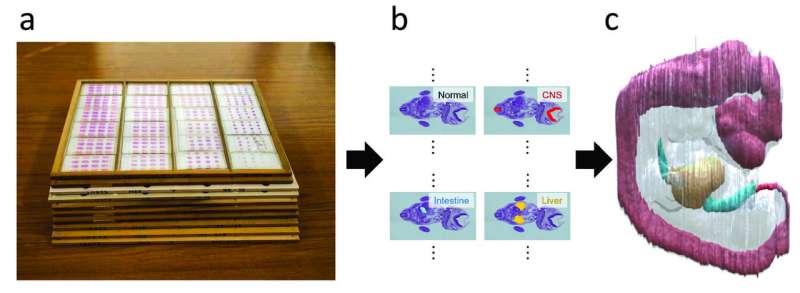
Japanese scientists report in Pattern Recognition a new method to construct 3-D models from 2-D images. The approach, which involves non-rigid registration with a blending of rigid transforms, overcomes several of the limitations in current methods. The researchers validate their method by applying it to the Kyoto Collection of Human Embryos and Fetuses, the largest collection of human embryos in the world, with over 45,000 specimens.
MRI and CT scans are standard techniques for acquiring 3-D images of the body. These modalities can trace with unprecedented precision the location of an injury or stroke. They can even reveal the microscopic protein deposits seen in brain pathologies like Alzheimer's disease. However, for the best resolution, scientists still depend on slices of the specimen, which is why cancer and other biopsies are taken. Once the information desired is acquired, scientists use algorithms that can put together the 2-D slices to recreate a simulated 3-D image. In this way, they can reconstruct an entire organ or even organism.
Stacking slices together to create a 3-D image is akin to putting a cake together after it has been cut. Yes, the general shape is there, but the knife will cause certain slices to break so that the reconstructed cake never looks as beautiful as the original. While this might not upset the party of five-year olds who want to indulge, the party of surgeons looking for the precise location of a tumor are harder to appease.
In fact, the specimen can undergo a series of changes when prepared for sectioning. "The sectioning process stretches, bends and tears the tissue. The staining process varies between samples. And the fixation process causes tissue destruction," explains Nara Institute of Science and Technology (NAIST), Nara, Japan, Associate Professor Takuya Funatomi, who led the project.
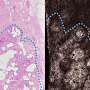
Fundamentally, there are three challenges that emerge with the 3-D reconstruction. First is non-rigid deformation, in which the position and orientation of various points in the original specimen have changed. Second is tissue discontinuity, where gaps may appear in the reconstruction if the registration fails. Finally, there is a scale change, where portions of the reconstruction are disproportional to their real size due to non-rigid registration.
For each of these problems, Funatomi and his research team proposed a solution that when combined resulted in a reconstruction that minimizes all three factors using less computational cost than standard methods.
"First, we represent non-rigid deformation using a small number of control points by blending rigid transforms," says Funatomi. The small number of control points can be estimated robustly against the staining variation.
"Then we select the target images according to the non-rigid registration results and apply scale adjustment," he continues.
The new method mainly focuses on a number of serial section images of human embryos from the Kyoto Collection of Human Embryos and Fetuses and could reconstruct 3-D embryos with extraordinary success.
Notably, there are no MRI or CT scans of the samples, meaning no 3-D models could be used as a reference for the 3-D reconstruction. Further, wide variability in tissue damage and staining complicated the reconstruction.
"Our method could describe complex deformation with a smaller number of control points and was robust to a variation of staining," says Funatomi.
More information: Takehiro Kajihara et al, Non-rigid registration of serial section images by blending transforms for 3D reconstruction, Pattern Recognition (2019). DOI: 10.1016/j.patcog.2019.07.001