Using machine learning to predict high-impact research
An artificial intelligence framework built by MIT researchers can give an "early-alert" signal for future high-impact technologies, by learning from patterns gleaned from previous scientific publications.
In a retrospective test of its capabilities, DELPHI, short for Dynamic Early-warning by Learning to Predict High Impact, was able to identify all pioneering papers on an experts' list of key seminal biotechnologies, sometimes as early as the first year after their publication.
James W. Weis, a research affiliate of the MIT Media Lab, and Joseph Jacobson, a professor of media arts and sciences and head of the Media Lab's Molecular Machines research group, also used DELPHI to highlight 50 recent scientific papers that they predict will be high impact by 2023. Topics covered by the papers include DNA nanorobots used for cancer treatment, high-energy density lithium-oxygen batteries, and chemical synthesis using deep neural networks, among others.
The researchers see DELPHI as a tool that can help humans better leverage funding for scientific research, identifying "diamond in the rough" technologies that might otherwise languish and offering a way for governments, philanthropies, and venture capital firms to more efficiently and productively support science.
"In essence, our algorithm functions by learning patterns from the history of science, and then pattern-matching on new publications to find early signals of high impact," says Weis. "By tracking the early spread of ideas, we can predict how likely they are to go viral or spread to the broader academic community in a meaningful way."
The paper has been published in Nature Biotechnology.
Searching for the "diamond in the rough"
The machine learning algorithm developed by Weis and Jacobson takes advantage of the vast amount of digital information that is now available with the exponential growth in scientific publication since the 1980s. But instead of using one-dimensional measures, such as the number of citations, to judge a publication's impact, DELPHI was trained on a full time-series network of journal article metadata to reveal higher-dimensional patterns in their spread across the scientific ecosystem.
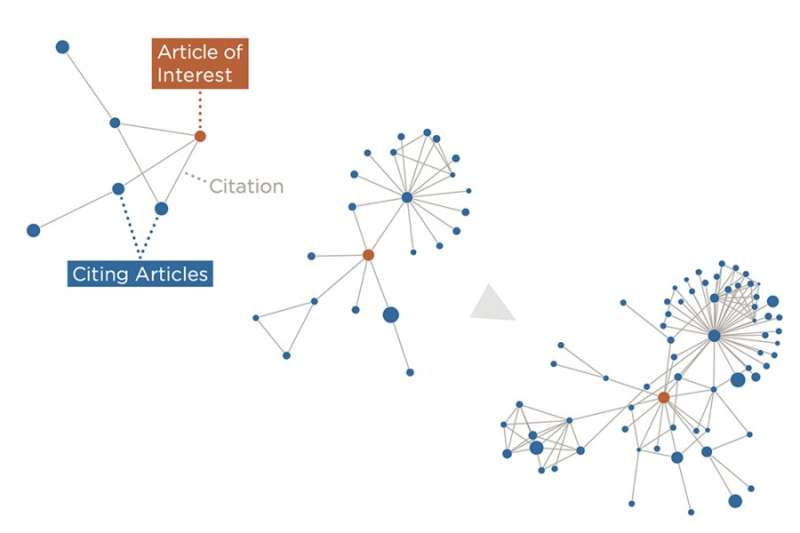
The result is a knowledge graph that contains the connections between nodes representing papers, authors, institutions, and other types of data. The strength and type of the complex connections between these nodes determine their properties, which are used in the framework. "These nodes and edges define a time-based graph that DELPHI uses to learn patterns that are predictive of high future impact," explains Weis.
Together, these network features are used to predict scientific impact, with papers that fall in the top 5 percent of time-scaled node centrality five years after publication considered the "highly impactful" target set that DELPHI aims to identify. These top 5 percent of papers constitute 35 percent of the total impact in the graph. DELPHI can also use cutoffs of the top 1, 10, and 15 percent of time-scaled node centrality, the authors say.
DELPHI suggests that highly impactful papers spread almost virally outside their disciplines and smaller scientific communities. Two papers can have the same number of citations, but highly impactful papers reach a broader and deeper audience. Low-impact papers, on the other hand, "aren't really being utilized and leveraged by an expanding group of people," says Weis.
The framework might be useful in "incentivizing teams of people to work together, even if they don't already know each other—perhaps by directing funding toward them to come together to work on important multidisciplinary problems," he adds.
Compared to citation number alone, DELPHI identifies over twice the number of highly impactful papers, including 60 percent of "hidden gems," or papers that would be missed by a citation threshold.
"Advancing fundamental research is about taking lots of shots on goal and then being able to quickly double down on the best of those ideas," says Jacobson. "This study was about seeing whether we could do that process in a more scaled way, by using the scientific community as a whole, as embedded in the academic graph, as well as being more inclusive in identifying high-impact research directions."
The researchers were surprised at how early in some cases the "alert signal" of a highly impactful paper shows up using DELPHI. "Within one year of publication we are already identifying hidden gems that will have significant impact later on," says Weis.
He cautions, however, that DELPHI isn't exactly predicting the future. "We're using machine learning to extract and quantify signals that are hidden in the dimensionality and dynamics of the data that already exist."
Fair, efficient, and effective funding
The hope, the researchers say, is that DELPHI will offer a less-biased way to evaluate a paper's impact, as other measures such as citations and journal impact factor number can be manipulated, as past studies have shown.
"We hope we can use this to find the most deserving research and researchers, regardless of what institutions they're affiliated with or how connected they are," Weis says.
As with all machine learning frameworks, however, designers and users should be alert to bias, he adds. "We need to constantly be aware of potential biases in our data and models. We want DELPHI to help find the best research in a less-biased way—so we need to be careful our models are not learning to predict future impact solely on the basis of sub-optimal metrics like h-Index, author citation count, or institutional affiliation."
DELPHI could be a powerful tool to help scientific funding become more efficient and effective, and perhaps be used to create new classes of financial products related to science investment.
"The emerging metascience of science funding is pointing toward the need for a portfolio approach to scientific investment," notes David Lang, executive director of the Experiment Foundation. "Weis and Jacobson have made a significant contribution to that understanding and, more importantly, its implementation with DELPHI."
It's something Weis has thought about a lot after his own experiences in launching venture capital funds and laboratory incubation facilities for biotechnology startups.
"I became increasingly cognizant that investors, including myself, were consistently looking for new companies in the same spots and with the same preconceptions," he says. "There's a giant wealth of highly-talented people and amazing technology that I started to glimpse, but that is often overlooked. I thought there must be a way to work in this space—and that machine learning could help us find and more effectively realize all this unmined potential."
More information: James W. Weis et al. Learning on knowledge graph dynamics provides an early warning of impactful research, Nature Biotechnology (2021). DOI: 10.1038/s41587-021-00907-6