This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Global digital volume correlation of large volumes: A sub-volume adaptive meshing approach
Digital Volume Correlation (DVC) is a powerful image analysis technique used in the field of materials science and engineering to study the mechanical behavior and deformation of complex 3D structures. By comparing voxel intensities in a pair of 3D digital images captured at different states of loading or deformation, DVC allows researchers to track and quantify displacements, strains, and other mechanical properties with high precision and non-invasively.
This non-destructive and quantitative approach has wide applications in studying a variety of materials, from biological tissues and composites to porous materials, providing valuable insights into their mechanical response under various conditions and contributing significantly to advancements in understanding materials' mechanical behavior for engineering and biomedical applications.
Addressing the limitations
DVC can be performed either in a local, or in a global fashion. In the local approach, a 3D image is typically divided into several sub-volumes and independent parameters for the displacement interpolation of each sub-volume are calculated. For each sub-volume, a matching subset in the subsequent 3D images is computed that maximizes their correlation. In the global approach, a globally shared set of parameters for the displacement interpolation over the entire 3D image is computed. Overall, the accuracy of DVC results is higher for the global approach than for the local approach, due to the continuity of the global displacement interpolation.
However, the main limitation to a more widespread use of global DVC is a hardware limitation, as significant computational power is needed to store and update the entire reference and deformed 3D images during the optimization process.
Forging a new approach
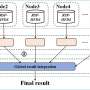
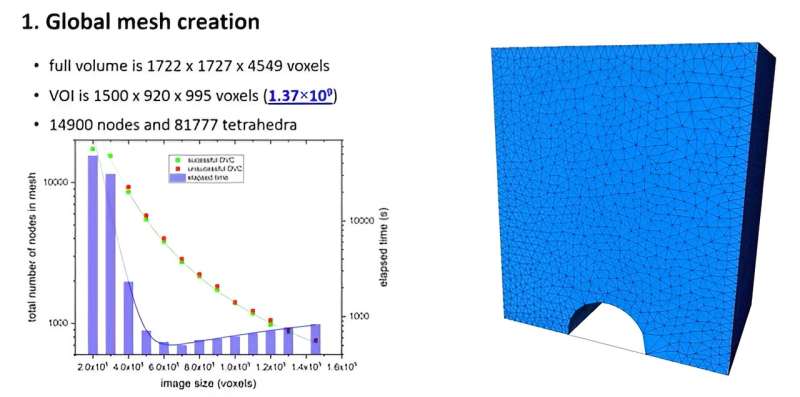
Dr. Fabien Leonard, University of Manchester at Harwell Data Analysis Manager, developed a solution based on a sub-volume adaptive meshing approach to perform global DVC on volumes larger than the size limit imposed by the hardware running the DVC algorithm.
The method relies on dividing the large meshed volume into smaller overlapping volumes, and corresponding meshes, that can be handled successively by the computer hardware to perform global DVC. Then, the nodes in the overlapping regions are assessed and the resulting sub-volumes global DVC results are merged into a single output result file covering the entire XCT volume.
Demonstrated on an in-situ testing of a graphite sample under compressive mechanical load with a volume of interest one order of magnitude and mesh size two order of magnitude greater than what is allowed by the computing hardware, it provides a solution to overcome hardware limitations in cases where both global DVC is required over large volumes and meshing density can be user-defined to fit the expected damage location within the sample.
The findings are published in the e-Journal of Nondestructive Testing.
More information: Global digital volume correlation of large volumes: a sub-volume adaptive meshing approach, e-Journal of Nondestructive Testing (2023). DOI: 10.58286/26629