August 21, 2019 feature
A web application to extract key information from journal articles

Academic papers often contain accounts of new breakthroughs and interesting theories related to a variety of fields. However, most of these articles are written using jargon and technical language that can only be understood by readers who are familiar with that particular area of study.
Non-expert readers are thus typically unable to understand scientific articles, unless they are curated and made more accessible by third parties who understand the concepts and ideas contained within them. With this in mind, a team of researchers at the Texas Advanced Computing Center in the University of Texas at Austin (TACC), Oregon State University (OSU) and the American Society of Plant Biologists (ASPB) have set out to develop a tool that can automatically extract important phrases and terminology from research papers in order to provide useful definitions and enhance their readability.
"Our project is motivated by the need of improving the readability of journal articles," Weijia Xu, who lead the team at TACC, told TechXplore. "It is a joint effort between biological curators, journal publishers and computer scientists aimed at developing a web service that can recognize and enable author curation of important terminology used in journal publications. The terminology and words are then attached to the end of the journal article in order to increase its accessibility for readers."
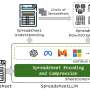
Xu and his colleagues developed an extensible framework that can be used to extract information from documents. They then implemented this framework within a web service called DIVE (Domain Information Vocabulary Extraction), integrating it with the journal publication pipeline of the ASPB. Unlike existing tools for extracting domain information, their framework combines several approaches, including ontology-guided extraction, rule-based extraction, natural language processing (NLP) and deep learning techniques.

"The results attained by different models are then stored in a centralized database," Xu explained. "We also designed a web service that allows users to curate extraction results. The web service is integrated with the production publication pipeline at ASPB."
Once the preview version of a journal article is submitted and enters the ASPB's pipeline, the manuscript is automatically fed to DIVE, which processes it and produces a URL with which the author will be able to access the processing results of DIVE. The author of the paper is asked to visit the link provided and review the extracted information before he/she is able to officially submit the paper.
"The author need to visit the DIVE site to review the extraction results and make final approval of the list of information to be included at end of their article," Xu said. "DIVE also tracks author corrections to improve future extraction tasks. Currently, no other journal publisher has adopted a similar approach and integrated it with their publication pipeline."
During its analyses and when extracting key data from documents, the framework developed by the researchers uses several techniques. This allows it to capture more information than other methods, such as ABNER (A Biomedical Named Entity Recognizer), which is an open source software tool for molecular biology text mining that can only extract general terms (e.g. genes and proteins). Contrarily to DIVE, ABNER is only based on conditional random fields (CRFs), a statistical modeling method that is commonly used in pattern recognition and machine learning applications.
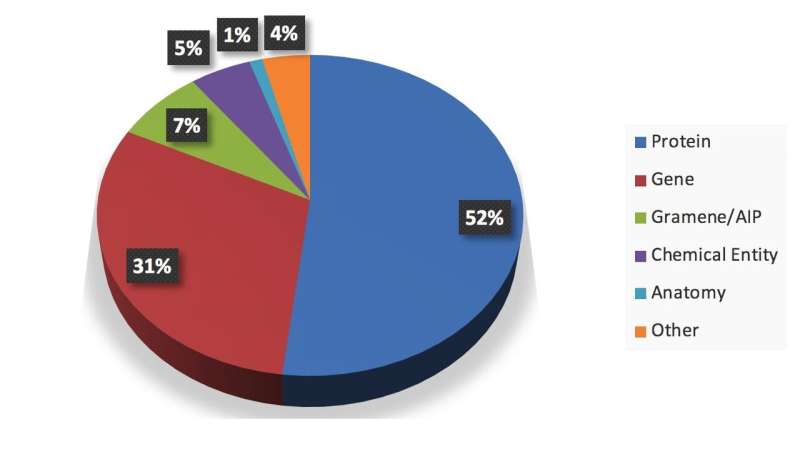
"A major contribution of our project is that it helps to build datasets and models that can infer authors research interests from their publications," Xu said. "Our project can benefit broader communities of biological researchers. For authors, the extractions and inclusion of the key information can increase the accessibility of their articles."
Xu and his colleague Amit Gupta evaluated their framework and compared its performance to that of other information extraction tools, including ABNER. Their findings revealed that using multiple approaches, including deep learning, DIVE attains higher-precision scores than other pre-trained models solely based on CRFs. Interestingly, the DIVE framework can also be continuously updated, as additional extraction models can be added to it at any time.
The DIVE web application does not only allow non-expert readers to better understand academic papers, it can also help them to identify papers aligned with their interests. Researchers, on the other hand, can use DIVE to stay informed about particular research areas, as well as to learn about new terminology and trends related to their field of interest. Finally, the information generated by the application can also guide biology curators in their decisions and data collection processes.
"We are continuing our project by exploring two directions," Xu said. "On one hand, we are investigating novel methods to incorporate with our information extraction models to improve the performance. On the other hand, we are also trying to expand our service by offering it to additional user communities and journal publishers."
More information: Amit Gupta et al. Extracting Domain Information using Deep Learning, Proceedings of the Practice and Experience in Advanced Research Computing on Rise of the Machines (learning) - PEARC '19 (2019). DOI: 10.1145/3332186.3332255
© 2019 Science X Network