January 17, 2020 weblog
AlphaFold makes its mark in predicting protein structures

Players applaud, say words like Whoo, bang plastic knives on the table and enjoy the best weekends with artificial intelligence as the main act, thanks to AI unleashed in games.
WIRED UK
's science editor, Matt Reynolds, looked at DeepMind's impact on AI milestones: "It has outplayed Go champions, bested professional StarCraft players and turned its attention to chess and shogi."
Let the games continue but the serious stuff must seriously shine. In brief, we can admire that unleashing AI for the purpose of scientific discovery has become especially alive and well thanks to research at DeepMind.
Tech watchers commented this week on research papers showing the strengths of AI. "As AI matures as a field (and runs out of video games to conquer) probably more of its achievements will look like these: solid improvements in important research domains."
A research paper published in Nature made instant news as a paper on how AI can be used to predict protein folding. VentureBeat referred to formidable challenges inching closer to goals.
The paper is "Improved protein structure prediction using potentials from deep learning," The article was published online on December 15 in Nature.
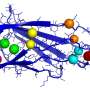
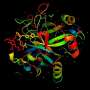
The DeepMind system is called AlphaFold. Reynolds had an interesting note about AlphaFold and the dataset: "The protein folding field is also well set up for training artificially intelligent agents. It has a large dataset—the Protein Data Bank, a repository of the 3-D structure and genetic makeup of 150,000 proteins, that was used to train DeepMind's protein structure-predicting system, called AlphaFold."
The paper explored DeepMind's achievements thus far in using AI to predict protein folding. The big deal about protein folding is that, as Kelsey Piper in Vox remarked, it is a "crucial issue for developing new drugs."
Three of the DeepMind team, Andrew Senior, John Jumper and Demis Hassabis, posted a Jan. 15 blog entry explaining protein structures, the protein folding challenge and what they wanted to achieve.
"As demonstrated by Levinthal's paradox, it would take longer than the age of the known universe to randomly enumerate all possible configurations of a typical protein before reaching the true 3-D structure–yet proteins themselves fold spontaneously, within milliseconds. Predicting how these chains will fold into the intricate 3-D structure of a protein is what's known as the 'protein folding problem'–a challenge that scientists have worked on for decades."
The team showed they could train a neural network to make accurate predictions "of the distances between pairs of residues, which convey more information about the structure than contact predictions."
They talked about optimization via "a simple gradient descent algorithm to generate structures without complex sampling procedures."
The big deal about protein bolding is that, said Piper, it is a "crucial issue for developing new drugs." How so? And, why are "shapes" so important? "The shape that proteins take predicts which other substances they'll interact with, so understanding protein folding is crucial for drug discovery and could even be used for developing new manufacturing processes," Piper said.
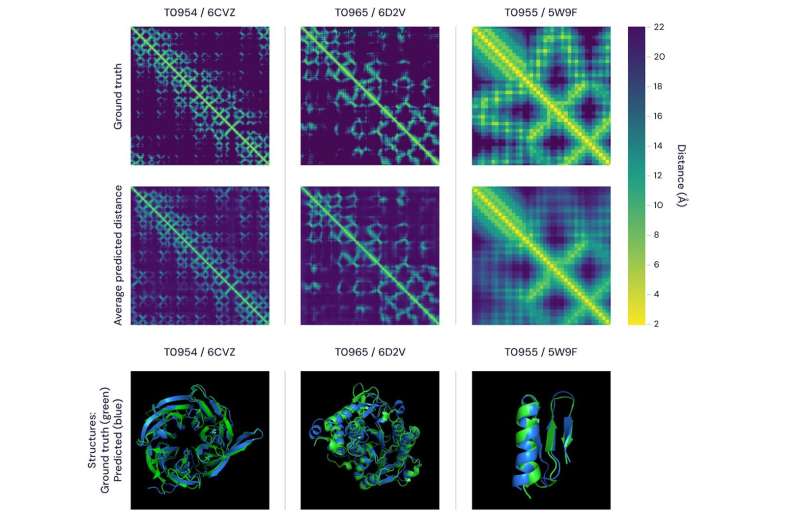
The DeepMind blog said that "a protein's form is thought to dictate its function. Once a protein's shape is understood, its role within the cell can be guessed at, and scientists can develop drugs that work with the protein's unique shape."
AlphaFold could help researchers get a better grasp of the function—and malfunction—of proteins.
A noteworthy quote in VentureBeat from the head of the UCL bioinformatics group David Jones, who advised the DeepMind team on parts of the project:
"Experimental techniques to determine protein structures are time-consuming and expensive, so there's a huge demand for better computer algorithms to calculate the structures of proteins directly from the gene sequences which encode them, and DeepMind's work on applying AI to this long-standing problem in molecular biology is a definite advance."
What kinds of experimental techniques have been time consuming and expensive? The blog mentioned cryo-electron microscopy, nuclear magnetic resonance and X-ray crystallography, each depending on trial and error, "which can take years of work, and cost tens or hundreds of thousands of dollars per protein structure."
Kyle Wiggers in VentureBeat: "With AlphaFold, DeepMind's research team focused on the problem of modeling target shapes from scratch without drawing on solved proteins as templates."
How Nature defines protein folding: "Protein folding is the process by which proteins achieve their mature functional (native) tertiary structure, and often begins co-translationally. Protein folding requires chaperones and often involves stepwise establishment of regular secondary and supersecondary structures, namely α-helices and β-sheets, that fold rapidly, stabilized by hydrogen bonding and disulphide bridges, and then tertiary structure."
Back in July, before the publication of the DeepMind paper, Nature had already taken note that "the race to crack one of biology's grandest challenges—predicting the 3-D structures of proteins from their amino-acid sequences—is intensifying, thanks to new artificial-intelligence (AI) approaches."
The DeepMind blog post said they made code available (GitHub), and offered the link, for anyone wanting to learn more or replicate their results.
In their blog post they stated why their system matters: "Our system, AlphaFold... builds on decades of prior research using large genomic datasets to predict protein structure. The 3-D models of proteins that AlphaFold generates are far more accurate than any that have come before."
Moving forward, the blog stated that a tool like AlphaFold might help rare disease researchers predict the shape of a protein of interest rapidly and economically and may eventually help them contribute to efficient drug discovery, while reducing costs associated with experimentation.
Rare-disease research is not the only area that can benefit from what they have accomplished. The blog talked about pollutants like plastic and oil; advances in biodegradable enzymes enabled by protein design could help break down waste in ways that are more friendly to our environment.
"In fact, researchers have already begun engineering bacteria to secrete proteins that will make waste biodegradable, and easier to process," according to the blog.
More information: Andrew W. Senior et al. Improved protein structure prediction using potentials from deep learning, Nature (2020). DOI: 10.1038/s41586-019-1923-7
© 2020 Science X Network