November 12, 2020 feature
Jet-printing complex circuits using microfluidics
Biomedical applications in life sciences can greatly benefit from microfluidics devices; however, the technology is suboptimal for rapid production and applications in biolabs. For instance, solid opaque walls of conventional microfluidic devices prevent biologists from providing adequate physical and optical access to their biological samples. Therefore, there is a growing need to engineer optimized microfluidics for efficient workflow. In a new study, Cristian Soitu and a research team at the Walsh and Cook Research Groups in the Department of Engineering Science and the Sir William Dunn School of Pathology at the University of Oxford, U.K., described a new contactless, microfluidics circuit fabrication method.
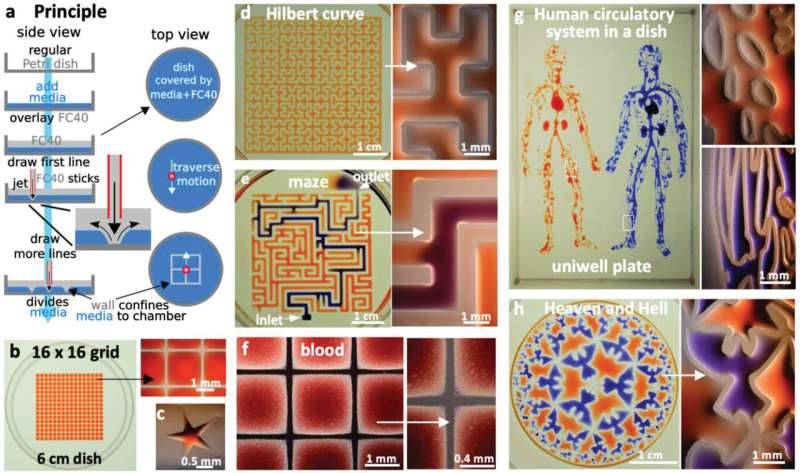
The team used standard petri dishes as the platform on which they developed the circuits using commonly available biolab tools, including a syringe pump, dispensing needles, cell culture media and an immiscible fluorocarbon (FC40) compound (a material freely permeable to oxygen and carbon dioxide). During the experiments, they "jetted" the FC40 from a dispensing needle through the medium present in the petri dish to reproducibly manufacture complex and highly accurate microfluidics structures in minutes. The technique allows the miniaturization of some common workflows in biology coupled with a flexible and diverse approach, well suited for widespread adoption in biomedicine. The work is now published on Advanced Science.
The new technique—jet-printing microfluidic devices on demand
The primary author of the study; bioengineer and Ph.D. researcher Cristian Soitu described the novelty of their frugal technique as "a contactless method to build fluid walls using materials commonly found in most biolabs." While detailing the speed and efficacy of their method, he added that "the fluid walls can be quickly built into any imaginable 2-D (two dimensional) circuit and we fabricated a human circulatory system in a microplate under 30 minutes, as a proof of concept." The new study is based on preceding work by the same research group who established 'freestyle fluidics' to use fluids as building blocks and confine liquids with fluid walls instead of solid walls.
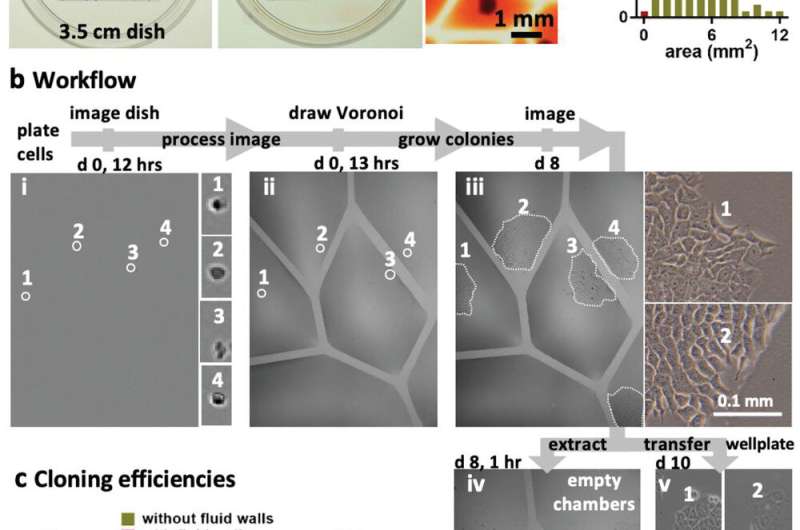
A key feature of the new approach is that it can overcome the Poisson limit during mammalian cell culture cloning. The Poisson statistics dictate that most cells cultured across wells in a microplate will be discarded after cell splitting and cloning protocols, thereby wasting cells and reagents. The jetting method will allow researchers to beat the Poisson limit by building isolated wells (known as Voronoi regions) to subculture the mammalian cells steadily in fresh media for a week, to harvest cell clones thereafter. Conventional open microfluidic devices too can be integrated into biomedical workflows, but they require substrate etching, surface treatment, contact or a combination of efforts to confine fluids and biological structures. Such complex manufacturing processes can deter biologists who favor efficient and flexible prototyping without compromising biocompatibility. As a result, the described jetting technique can provide a new hands-on approach to overcome existing limits of microfluidics.
The experimental approach
During the experiments, the researchers covered the bottom of a standard tissue-culture dish with a film of cell growth media and an overlay of FC40 to prevent evaporation. By moving the microjet sideways, they created a grid with 256 chambers in under two minutes on the FC40 layer formed on the petri dish. In this contactless method, the nozzle did not touch the dish or media, preventing cross-contamination. Conventional flow systems are highly specialized and built in, and are therefore difficult to integrate into existing workflows. Comparatively, the perfusion circuit jetted within a standard petri dish here, allowed perfusion within 15 minutes to feed myoblasts (embryonic precursors of muscle cells) to subsequently generate myotubes (muscle fibers formed via myoblast fusion) within a week. Soitu et al. additionally highlight the ability to sub-culture cells without using trypsin/EDTA (conventional reagents), given that a whole or part of the colonies will be dislodged by the FC40 jetting process to transfer to new chambers. The method therefore allowed them to sample a single colony several times as it grew.
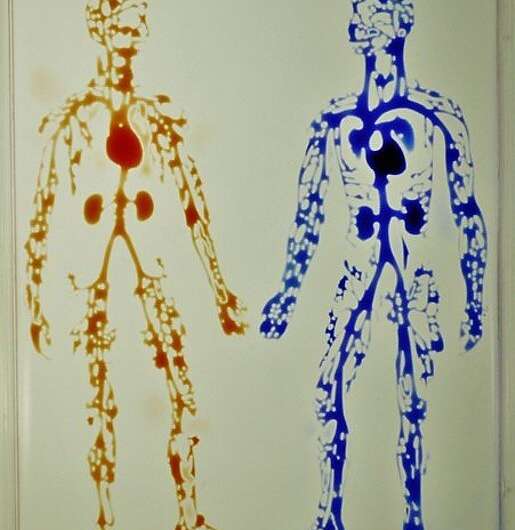
Reproducibility of the method
To demonstrate the reproducibility of the method, the team used jetting to build walls in the form of a star-shaped chamber, as a square enclosing a single continuous wall shaped like a Hilbert curve and a microfluidic maze with blue dye inserted to solve a maze and find the exit. The method is not restricted to the prescribed media and can be replaced with whole human blood for specific experiments. The patterns themselves include the "the human circulatory system" and the "circle limit IV" (Heaven and Hell) by M.C. Escher (WikiArt), which are diverse and flexible. The efforts highlight the potential to reproduce almost any pre-existing pattern. The resulting fluid walls were sufficiently stable; therefore, the researchers could carry the constructs between labs, incubators and microscopes, much like any dish filled with liquid.
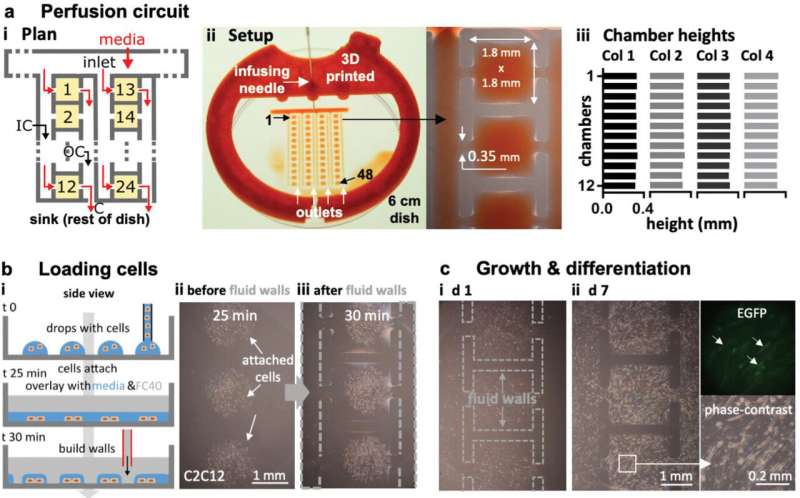
Building a complex perfusion circuit with constant flow
The team then engineered a complex circuit via jet printing to provide a steady flow of fresh media for seven days to maintain cell growth in an array of chambers. To accomplish this, they designed a circuit where each chamber received fresh media and demonstrated circuit functionality using mouse myoblasts that differentiated into mature myotubes within seven days. If required, the team can disconnect any chamber from the adjoining circuit by building new FC40 walls.
In this way, Cristian Soitu and colleagues developed a rapid and precise technique to generate microfluidic circuits on a standard polystyrene petri dish. They projected a microjet of PC40 to the bottom of the dish to build a liquid wall pinned by interfacial forces. The team proved the ability to (1) build stable fluid circuits with any imaginable 2-D shape within seconds to minutes, (2) drive flow via conduits without using external pumps, and (3) create valves that can be opened and closed at will. Key features of the approach will allow biologists to beat the Poisson limit, generate a robust perfusion system and subculture attached cells without conventional reagents (such as trypsin/EDTA). The research team will optimize the device and overcome existing limits to construct ever-smaller fluid circuits capable of retaining miniaturized volumes of media. The diversity and flexibility of this approach is expected to lead to widespread adoption of the technique for a variety of applications in biomedicine.
More information: Cristian Soitu et al. Jet‐Printing Microfluidic Devices on Demand, Advanced Science (2020). DOI: 10.1002/advs.202001854
Eric K. Sackmann et al. The present and future role of microfluidics in biomedical research, Nature (2014). DOI: 10.1038/nature13118
Cristian Soitu et al. Microfluidic chambers using fluid walls for cell biology, Proceedings of the National Academy of Sciences (2018). DOI: 10.1073/pnas.1805449115
© 2020 Science X Network