October 31, 2019 feature
Modeling and simulating complex dynamic musculoskeletal architectures

Life scientists and bioengineers study natural systems and organisms to understand their biophysical mechanisms, in order to drive biomimetic engineering applications in the lab. In a recent report on Nature Communications, Xiaotian Zhang and colleagues in the Departments of Mechanical Engineering and Supercomputing Applications in the U.S. presented a numerical approach to simulate musculoskeletal architectures. The method relied on the assembly of heterogenous active and passive Cosserat rods (a deformable-directed rod based curve) into dynamic structures to model bones, tendons, ligaments, fibers and muscle connectivity. They demonstrated applications of the technique by solving a range of problems in biological and soft robotic scenarios across many environments and scales. The team engineered bio-hybrid robots at the millimeter-scale and reconstructed complex musculoskeletal systems. The method is versatile and offers a framework to assist forward and reverse bioengineering designs for fundamental discoveries on the functions of living organisms.
Musculoskeletal systems contain bones, muscles, tendons and ligaments to function together in native biological systems. Intriguingly, such architectures contain an inseparable nexus between actuation and control, topology and mechanics due to nonlinear constituents. In stark contrast with rigid-body robots, soft biological creatures can implement deformations and structural instabilities to function within complex, unstructured and dynamic environments. Biological musculoskeletal architectures possess an intrinsic distributedness, softness and compliance to outsource control tasks to a variety of compartments. This property provides the basis for an emerging paradigm in biologically inspired robotics known as morphological computation or mechanical intelligence. The considerations have led to a variety of experimental advances in soft robotics with artificial compliant materials to form soft bio-hybrid robots. Despite experimental advances in the field, efforts to model and simulate dynamic musculoskeletal architectures across biological and synthetic structures remain to be accomplished.
Simulating the human elbow joint
Zhang et al. first demonstrated a method to include complete dynamics of deformation such as bend, twist, shear and stretch involved with biological architectures. For this, they built on previous work on cosserat rods to establish a musculoskeletal modeling approach to realistically simulate active, heterogeneous biological layouts in a stepwise approach to synthesize and replicate living architectures with biological layers of complexity for advanced biomimetic applications. The work allowed them to replicate biological systems by mimicking the underlying biomechanics and provided access to study and understand biophysical functions of biological organisms in silicon. The study demonstrated the applications of rod models to play a valuable role while modeling complex active systems for biomimetic architectures.
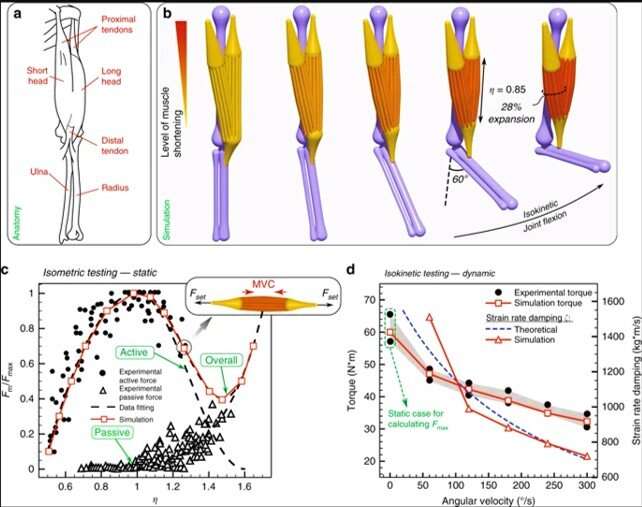
During the first biomimetic simulation, Zhang et al. used a human elbow joint with muscles, tendons and bones to illustrate rod assemblies mapped to physiology, dynamics and morphology. For example, the elbow joint showed soft and stiff characteristics, simplified dynamics and reduced configuration space. The analysis allowed them to verify and calibrate the model based on readily available anatomical and biomechanical data to relate to the Hill model. The representation's level of detail allowed them to address human patient-specific kinesiological needs and virtually reconstruct a 3-D replica of a human elbow joint using isometric and isokinetic tests applicable for bone tissue engineering. The scientists could model, calibrate and validate an individual muscle unit's actuation, where the result reproduced dynamics and morphology of a biological system. The current modeling approach presented key features setting it apart from the Hill model, to include (a) individual rods for selective recruitment to mimic an injury, and (b) compliant muscles that can bend, twist and shear realistically to dynamic forces across the entire structure or within the environment.
Engineering bio-hybrid robots
In the next experiment, the team engineered bio-hybrid robots using their computational solver to guide the design and fabrication of swimming and walking bio-hybrid bots at the millimeter scale. To investigate swimming behavior of the robots, they first solved problems using numerical modeling and simulations of a previously established bio-hybrid flagella. For this, they created an exact replica of the original swimmer and modeled the polydimethylsiloxane (PDMS) substrate to mimic experimental geometry and material properties with living clustered cells modeled as a small, soft and contractile filament connected to the substrate. The qualitative and quantitative observations of swimming motion between the simulation and experiment matched well.
![Bio-hybrid robotic design. (a) One-to-one comparison of the robot with experimental photographs at different stages within one swimming cycle. (b) Axial position of the robot’s center monitored over more than 20 cycles compared with experimental data. (c) Optimization course: Convergence to optimal solution is observed after 48 generations. Optimization was constrained with head length within [0,1.927]mm, head radius within [4,40]μm, tail radius within [4,6.5]μm and cell location at any point along the tail. The bot longitudinal dimension is fixed at L=1.927mm, so that the tail length can be inferred from the head length. The parameter ranges are determined to account for actual manufacturability. (d) Visualization of both the original and the optimal designs showing configurations at rest and midline kinematic envelopes over one period. Original swimmer details: substrate is modeled with 424-μm-long head and 1503-μm-long tail with radii 20μm and 7μm respectively. Density ρ=0.965gcm−3, Young’s modulus E=3.86MPa, bending stiffness EI=2.427×10−9Nmm2, mass m=7.364×10−7g are set. The contractile cell is modeled with radius 10μm and length 100μm. The cell is set to produce a contracting force F=σmA=12μN with beating frequency f=3.6Hz23. The fluid has dynamic viscosity μ=1.2×10−3Pa⋅s. Optimized swimmer details: substrate has length and radius of 190μm and 32.3μm, respectively. The contractile cell is attached 190μm away from the head and the tail is 4.3μm thick. (e) Overall design of the walker with yellow elements representing muscle rings and purple elements representing the skeleton. Experimental images adapted from previous studies. (f) Simulations versus experiments: Bot displacement over 2 seconds for the actuation frequency 1 Hz. (g) Visualization of initial and optimized design of the walker. (h) Optimization course converges after 25 generations. Optimization was constrained with skeleton's Young's modulus [250–350] kPa, length of the shorter pillar [2.4–3.4] mm and location of muscle strip [0.5–3] mm (distance from ground), all of which are chosen according to manufacturability constraints. (i) Dynamic behavior of simulated (solid lines) and experimental (markers with error bars) walker with muscle contraction at different frequencies, and comparison of walking performance between initial (solid lines) and further optimized design (dashed lines). Credit: Nature Communications, doi: 10.1038/s41467-019-12759-5 Modeling and simulating complex dynamic musculoskeletal architectures](https://scx1.b-cdn.net/csz/news/800a/2019/1-modelingands.jpg)
After modeling and optimizing the bio-hybrid swimmer, they attempted to computationally design a bio-hybrid walker as developed previously to form the fastest motile biological machine to date by Pagan-Diaz et al., based on a preceding bio-hybrid robot. Structurally, the Pagan-Diaz model contained an asymmetric hydrogel scaffold and skeletal muscle tissues to resemble muscle-tendon-bone relationships in vivo and operated in a solution bath in the lab. Experimentally, they suspended the muscles and electrically shocked them to induce contraction for motion via asymmetry and friction.
Zhang et al. simulated this architecture in the present work, to design a new scaffold and topological muscle arrangements of the bot. The new muscle-tissue topology contained a thin strip section connecting two rings wrapped around skeleton legs to transfer muscle contraction forces, which they tested using benchmark studies. Zhang et al. experimentally demonstrated the computation blueprint with the Pagan-Diaz model to construct a new bio-bot with twice the speed of the original bio-hybrid robot; with good agreement observed in the simulation. The research team showed potential of the computational approach to encapsulate physics of cell- and muscle-powered soft robotic systems for the desired applications to engineer more flexible prototypes.
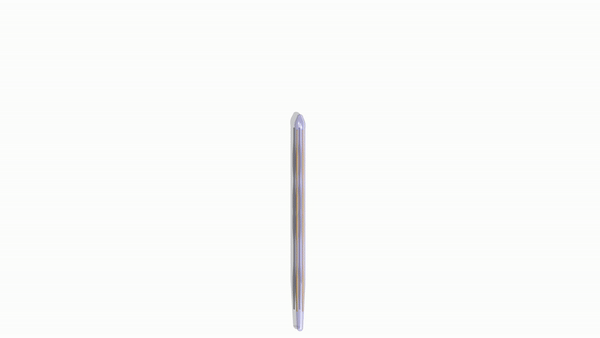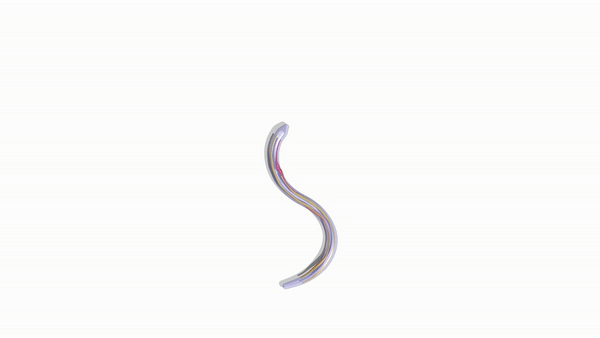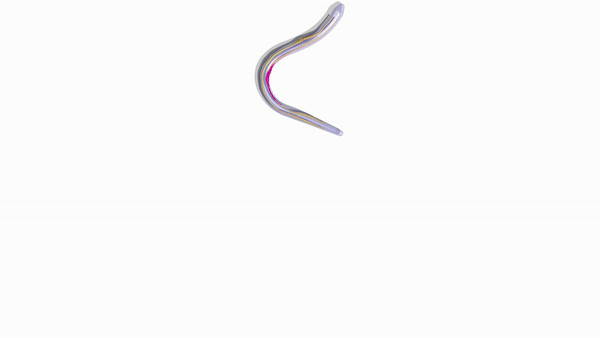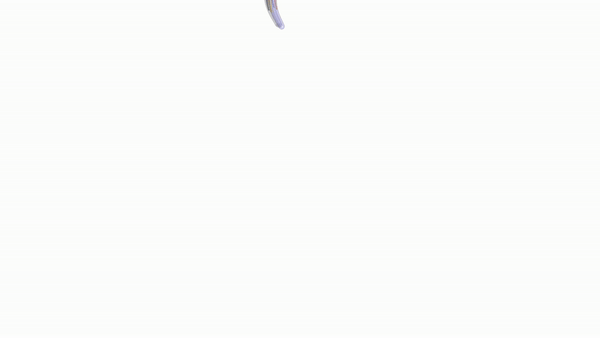
Engineering slithering snakes
The team then employed the numerical approach devised within the study to facilitate bio-hybrid robot manufacturability and understand biomechanics. Bioengineers had previously conducted multiple experiments on snake locomotion to build target robot replicas using servomotors (rotary actuators). In the present experiment, they developed a completely soft elastic snake bioinspired by real snakes but effectively actuated with several muscle-tendon groups to achieve smooth undulatory movement. The research team showed that while biological snakes had multiple muscles to orchestrate gaits and body deformations, the model only required few features to smoothly and effectively slither forward.
For optimal design, the scientists coupled their computational solver with the Covariance Matrix Adaptation-Evolution Strategy algorithm (CMA-ES) to identify the locations and actuation patterns for maximum speed forward. Zhang et al. compared and verified the results with reference simulations and experimental recordings. The team primarily aimed to reveal hidden architectural design principles and expose their function for engineering purposes. They carefully orchestrated distributed actuation for smooth realistic gaits in stark contrast to rigid snake robot counterparts. The researchers also implemented a framework to simplify, test and use biomechanical principles of complex biological systems to create a realistically slithering, fast, soft-robotic snake based on a few simple actuators.

Engineering feathered wings
To demonstrate a more complex strategy of locomotion that includes additional biological structures with critical functions, the team developed a feathered, musculoskeletal bird wing in silico. Bioengineers had previously conducted a variety of studies to understand biophysical features of bird flight from muscular activation patterns to biomechanics of feathers. In the present work, Zhang et al. considered the dynamic wing structure of a pigeon and reconstructed the feathers in silico with bending stiffness, consistent with previous studies.
In total, they connected 19 feathers to the computational wing model conformed to homing pigeon biological data. They included four muscles associated with the shoulder and elbow joints to control wing actuation and morphing relative to the human elbow joint model. Although this first-in-study model did not capture complex aerodynamics associated with flapping flight, it provided a preliminary estimate. The team reproduced the kinematics of morphing wings during takeoff and based the muscle actuation patterns on previously acquired experimentally recorded electromyography (EMG) signals.
In this way, Xiaotian Zhang and a team of researchers presented a new method to assemble heterogenous, active and passive cosserat rods to simulate dynamic, musculoskeletal architectures that can undergo deformation to facilitate biomimetic movement. The approach addressed an existing lack of engineering techniques in soft robotics to fill the gap between rigid-body modeling and high-fidelity finite-element method (FEM) simulations.
Using favorable features developed in the study, the research team engineered soft-bodied systems to resolve a number of problems relative to soft robotics and complex biological structures in diverse environments. The new work showed the versatility of the approach to establish a promising strategy for broad ranging applications across bioengineering biologically inspired structures to discovering the underlying features of living organisms.
More information: Xiaotian Zhang et al. Modeling and simulation of complex dynamic musculoskeletal architectures, Nature Communications (2019). DOI: 10.1038/s41467-019-12759-5
Ritu Raman et al. Optogenetic skeletal muscle-powered adaptive biological machines, Proceedings of the National Academy of Sciences (2016). DOI: 10.1073/pnas.1516139113
Gelson J. Pagan-Diaz et al. Simulation and Fabrication of Stronger, Larger, and Faster Walking Biohybrid Machines, Advanced Functional Materials (2018). DOI: 10.1002/adfm.201801145
© 2019 Science X Network