May 16, 2022 feature
A weakly supervised machine learning model to extract features from microscopy images
Deep learning models have proved to be highly promising tools for analyzing large numbers of images. Over the past decade or so, they have thus been introduced in a variety of settings, including research laboratories.
In the field of biology, deep learning models could potentially facilitate the quantitative analysis of microscopy images, allowing researchers to extract meaningful information from these images and interpret their observations. Training models to do this, however, can be very challenging, as it often requires the extraction of features (i.e., number of cells, area of cells, etc.) from microscopy images and the manual annotation of training data.
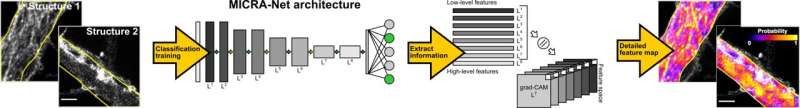
Researchers at CERVO Brain Research Center, the Institute for Intelligence and Data, and Université Laval in Canada have recently developed an artificial neural network that could perform in-depth analyses of microscopy images using simpler, image-level annotations. This model, dubbed MICRA-Net (MICRoscopy Analysis neural network), was introduced in a paper published in Nature Machine Intelligence.
"Manually extracting features from images is a long and tedious task, particularly in instances where it needs to be performed by a trained expert," Anthony Bilodeau, a Ph.D. student at Université Laval who carried out the study, told TechXplore. "While deep learning (DL) models for feature extraction are available, they still require training with annotations, which are often hard to obtain. Our model (MICRA-Net) relies on a simple classification task, asking the question: is the structure present in the region of the image that you are looking at or not?"
By addressing this simple question, the model developed by the team at Université Laval can predict the presence or absence of a specific structure in images using simple binary annotations. This greatly reduces the time required to annotate images and simplifies the training process, while still allowing the model to tackle multiple microscopy image analysis tasks simultaneously.
"Our model's weak supervision stems from the way MICRA-Net is trained," Bilodeau said. "The annotations required to train MICRA-Net are simple binary (yes or no) classification labels, which are much easier to obtain than complex precise labels, such as contours of the structure of interest."
In contrast with other existing deep learning tools for the analysis of microscopy images, MICRA-Net can tackle complex tasks, such as semantic segmentation and detection, but using far simpler, binary image annotations. It achieves this by extracting essential information about the structure of interest from the gradient class activated maps (i.e., grad-CAMs).
"Combining the grad-CAMs of multiple layers of the network allows the model to highlight the structure of interest in the image and can be used to generate precise segmentation masks or to localize the objects," Bilodeau explained. "MICRA-Net also achieves similar or better performance on complex image analysis tasks compared to established baselines trained using weak supervision (e.g., bounding box annotations, scribbles)."
In the initial evaluations carried out by the team at Université Laval, MICRA-Net achieved remarkable results, outperforming most of the models it was compared to. In the future, it could thus be used by research teams worldwide to tackle complex image analysis problems and discover crucial patterns in microscopy images.
"While some image analysis tasks can benefit from large and precisely annotated publicly available datasets for pre-training (e.g., nucleus segmentation) we believe that MICRA-Net should be considered for datasets for which no precise annotations are readily available or can be obtained easily," Bilodeau added. "For future research we plan on testing MICRA-Net on other challenging datasets and also improve the performance by investigating how other approaches can be combined for feature extraction."
More information: Anthony Bilodeau et al, Microscopy analysis neural network to solve detection, enumeration and segmentation from image-level annotations, Nature Machine Intelligence (2022). DOI: 10.1038/s42256-022-00472-w
© 2022 Science X Network